08_Identify_variations
By Yan Li
PhD in Bioinformatics, University of Liverpool
Why look at variation
- allows us to study evolution
- phylogenetics
- population genomics
- let us make genotype / phenotype associations
- genome wide association studies (GWAS)
- human disease, agriculture, genetic engineering
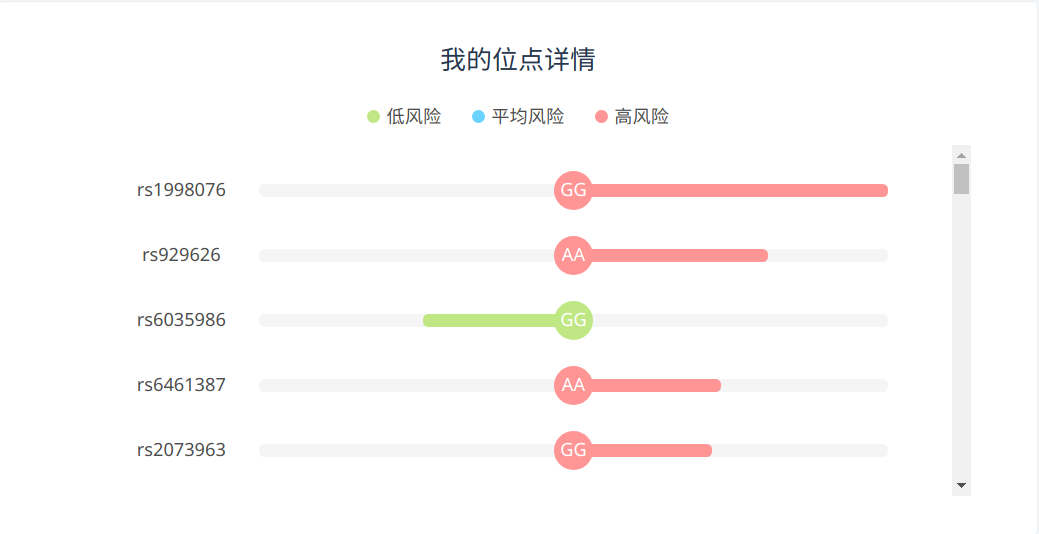
The personal gene test service
What is variation
- Single Nucleotide Polymorphisms (SNPs)
- a genetic "typo" of one nucleotide (e.g. A > G)
- INsertion / DELetions (INDELs)
- a string of one or more nucleotides that has been added/removed from a location in a genome (typically 1-100bp)
- Structural Variants (SVs)
- a region of DNA that has been inverted / translocated / duplicated (typically >100bp)
- Mobile Genetic Elements (MGEs)
- insertion / replication of retrotransposons, transposons, integrons etc.
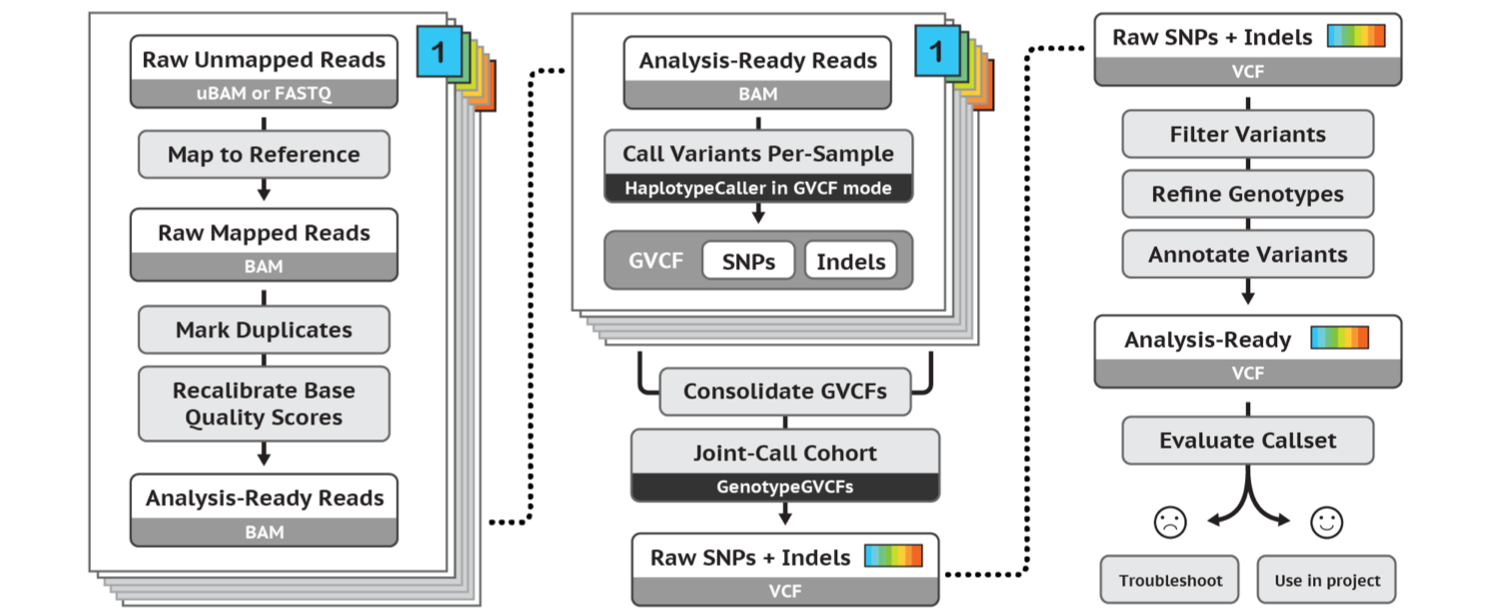
Shors reads allignment
- bwa (Burrows-Wheeler Aligner)
- Bowtie
Sam/Bam files
- SAM file is a TAB-delimited, line-oriented text format, including a header section and an alignment section
- Header section: each line contains some metadata
- Alignment section: each line contains the alignment of a read
- The SAM tag specification
- BAM file is the compressed format of SAM file
An example of SAM file
@HD VN:1.6 SO:coordinate
@SQ SN:NC_003197.2 LN:4857450
@SQ SN:NC_003277.2 LN:93933
@RG ID:foo PL:illumina SM:SRR1056117
SRR10561173.6336 2209 NC_003197.2 1 60 57H44M = 549 649 AGAGATTACGTCGGGTTGCAAGAGATCTTGACAGGGGGAATTGG .G...G...GAA.<A.<A.GGGAGAGA.<..G...<GAA.GGAG SA:Z:NC_003197.2,4857394,+,57M44S,60,0; MC:Z:101M MD:Z:12T14A16 PG:Z:MarkDuplicates RG:Z:foo NM:i:2 AS:i:34 XS:i:0
SRR10561173.114060 163 NC_003197.2 1 60 45S56M = 178 275 GAAAAAAAACTAACAAAATAACGTGCTGTAATTTTTAAAATAATAAGAGATTACGTCTGGTTGCACGAGATCATGACAGGGGGAATTGGTTGAAAATAAAT GGAA<<<A.<<AGGGGAAGAGG.<<G.<<<G.GGAA..<GGA..<...AA.<AGA<<AG.GGGAG.<.<.<.<<GG.GAGG...G.<G.....<<.<.GGG SA:Z:NC_003197.2,4857406,+,45M56S,60,0; MC:Z:98M MD:Z:20A35 PG:Z:MarkDuplicates RG:Z:foo NM:i:1 AS:i:51 XS:i:0
Call variants
- bcftools
- FreeBayes
- GATK
- GATK v3.8 and v4.0 have some differences.
- v4.0 merged the
picardtools
- VarScan2
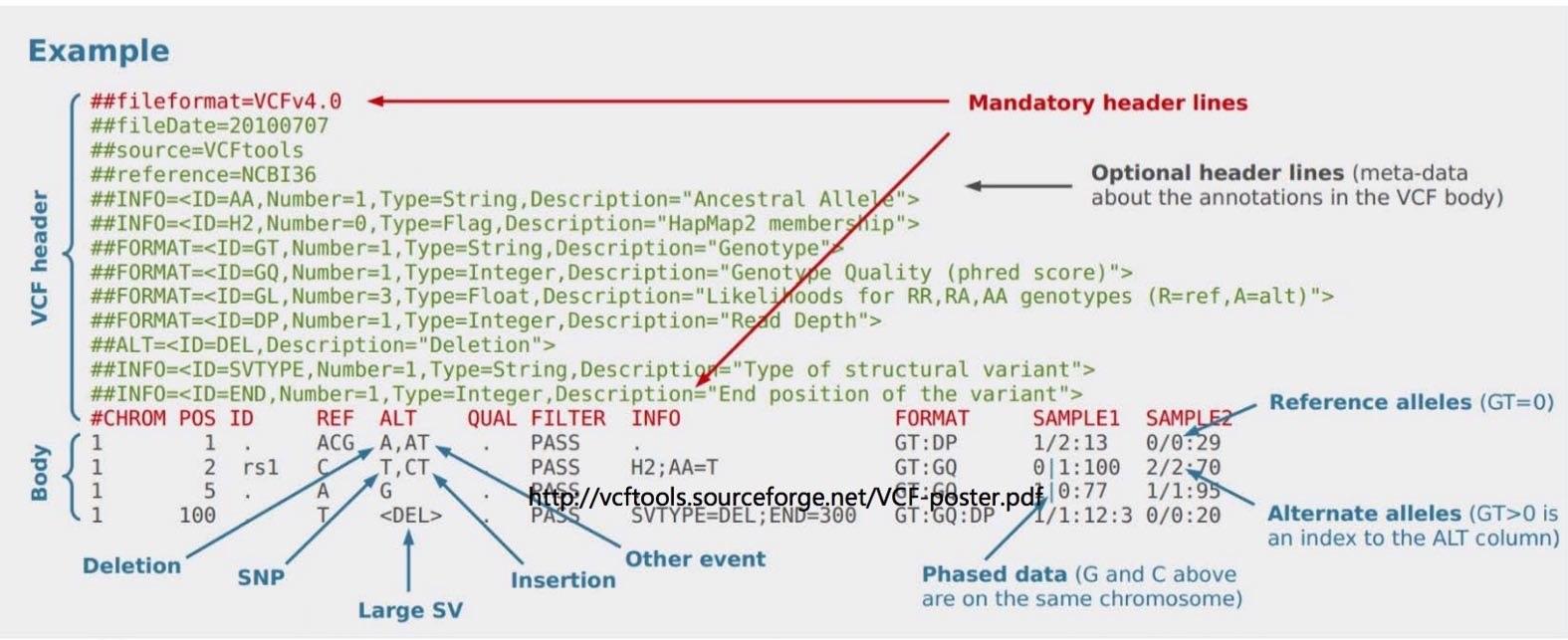
VCF files
Annotate SNPs
snpeffAnnovarVAAST 2