07_Genome_assembly
By Yan Li
PhD in Bioinformatics, University of Liverpool
Introduction
- Assembly: from short reads to long contigs
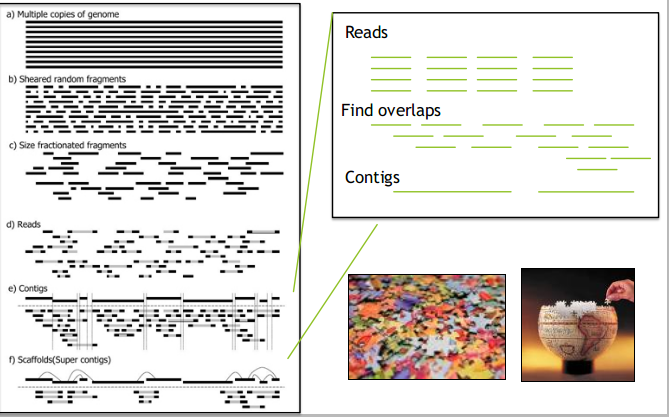
- Two types of genome assembly:
- de novo
- re-sequencing (reference-guided)
- We will focus on de novo of bacterial genome
Why
- Make a reference genome (when we are not already have one)
- Look at genome structure
- Put features into context
- To make comparison to other genomes
Technology choice
- Different assembler based on the sequencing platform
- PacBio & Nanopore
- Flye
- Illumina
- Spades
- Velvet
Factors dictate assemly quality
- Read length and coverage
- Sequencing data quality
- Genome complexity
Coverage / Depth
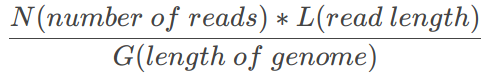
- Usually expressed as 30x, 100x, etc
- Low coverage cause some genome regions have no reads
- Short reads length may make repeat regions impossible to recover
\frac{N(number\space of\space reads) * L (read\space length)}
https:://katex.org/#demo
Terminology
| Term | Definition |
|---|---|
| assembly | reconstructing a genome sequence from raw reads |
| read | fragments of genome generated by a sequencer |
| coverage | the average number of reads that align to known reference bases |
| contig | a contigious sequence built from overlapping reads |
| scaffold | sets of overlapping contigs separated by gaps of known length |
| graph | represents relationships using nodes and edges |
Graph theory: the Seven Bridges of Königsberg
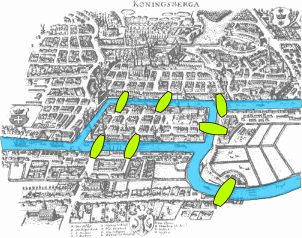
Can we visit each part of the city by crossing each bridge once?
Graph theory: the Seven Bridges of Königsberg

- Eulerian path = visit every edge of the graph only once
- In this problem it's impossible
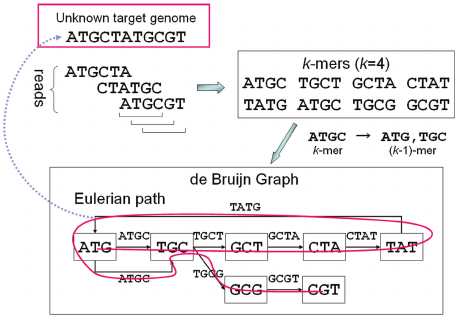
de Bruijn graph
- Reads are broken into k-mers (substrings of length k)
- A de Bruijn graph is is constructed from the k-mers: k-mers are connected if they have k-1 shared bases
- The genome is derived using the Eulerian path through the graph
- E.G. assemblers: SPAdes, Velvet, ABySS
de Bruijn graph assembler
Graph features
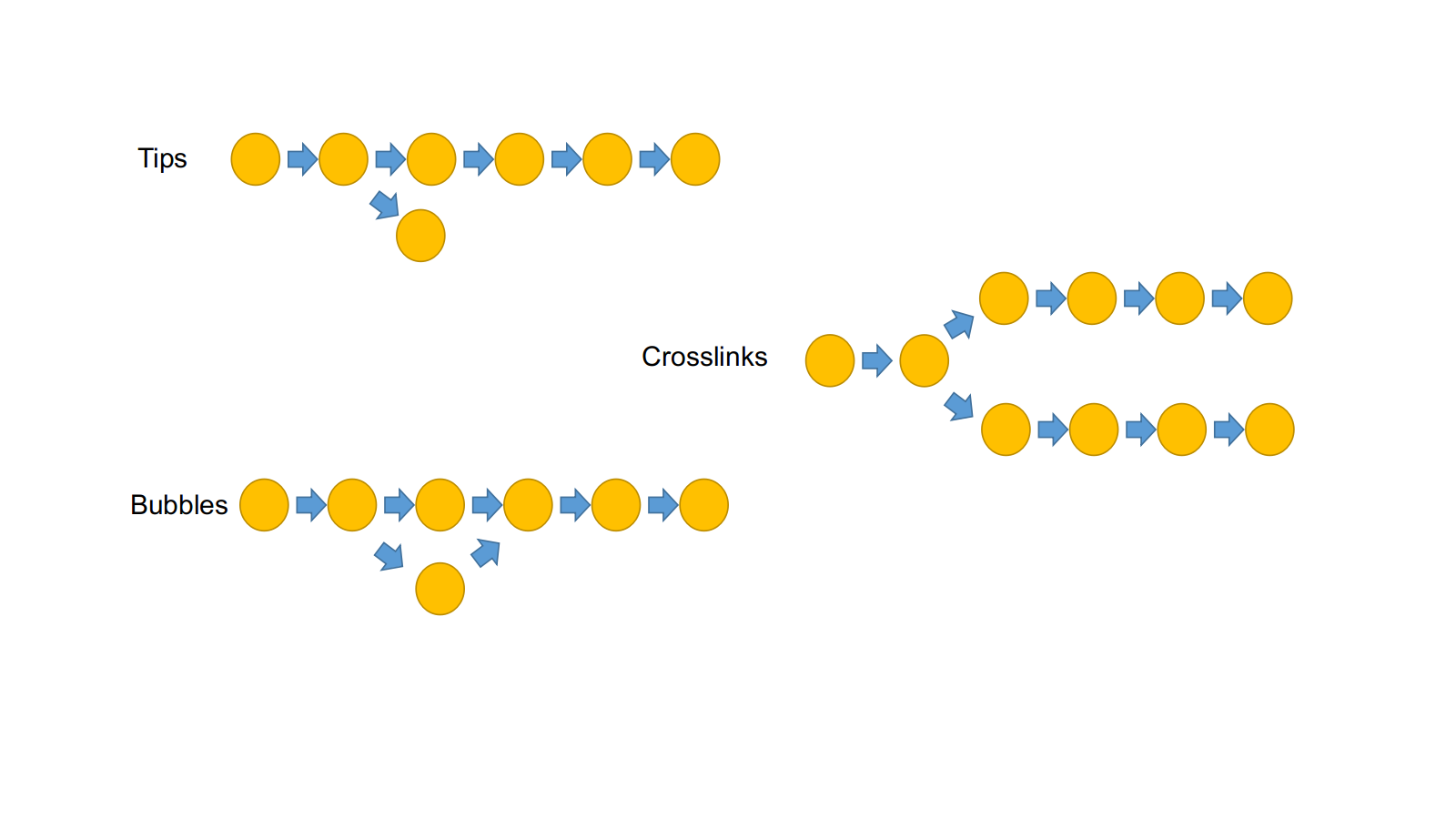
Use k-mer frequency to resolve these graph features::
- remove low depth kmers
- clip tips, merge bubbles, remove links
- resolve small repeats using long kmers
K-mer size
- avoid using an even numbered k-mer size
- they can lead to reverse complementing
- affects the strand specificity of the graph
- palindromic k-mers are avoided with an odd k
- inceasing k-mer size can resolve ambiguities
- higher k-mer size can < number of edges and < possible paths
- however higher k-mer size also more sensitive to sequencing errors
- higher k-mer size means more RAM needed
- try several k-mer sizes to get the best assembly
Assembly quality
We assess quality by looking at the assembly contiguity, completeness and correctness
Contiguity
- Ideally, we want a single complete chromosome
- We measure contiguity using::
- contig number
- contig length (average, median and maximum)
- N statistics (e.g. N50)
- N50 is a statistical measure of the average length of a set of contigs
- 50% of the entire assembly is contained in contigs with length >= the N50 value
Completeness and correctness
- Completeness = assembled genome size / estimated genomes size
- Correctness is a measure of the number of errors in the assembly
- feature compressions (i.e. repeats)
- improper contig scaffolding
- introduced SNPs/InDels