05_Sequencing_and_FastQ
By Yan Li
PhD in Bioinformatics, University of Liverpool
Workflow
graph TB
A(DNA extraction)
A --> B
B(Sequencing)
B --> C
C(Trimming and QC)
C --> D
D(Assembly)
D --> E
E(Annotation or Other analysis)
Sequencing: an overview
| Gene Sqeuencing Development | The Key Players | Key Technology | Key Product | Product Release Time |
|---|---|---|---|---|
| Sanger's Sequencing | ABI | Chain Termination Method | ABI 3730 | 1987 |
| Next Generation Sequencing (NGS) | Illumina | Sequencing by Synthesis | Hiseq, Miseq | 2006 |
| Third Generation Sequencing | Pacific Biosciences | SMRT Technology | PacBio RS, PacBio RS II | 2013 |
| Oxford Nanopore Technology | Nanopore Technology | MinION | 2014 |
Sequencing: an overview
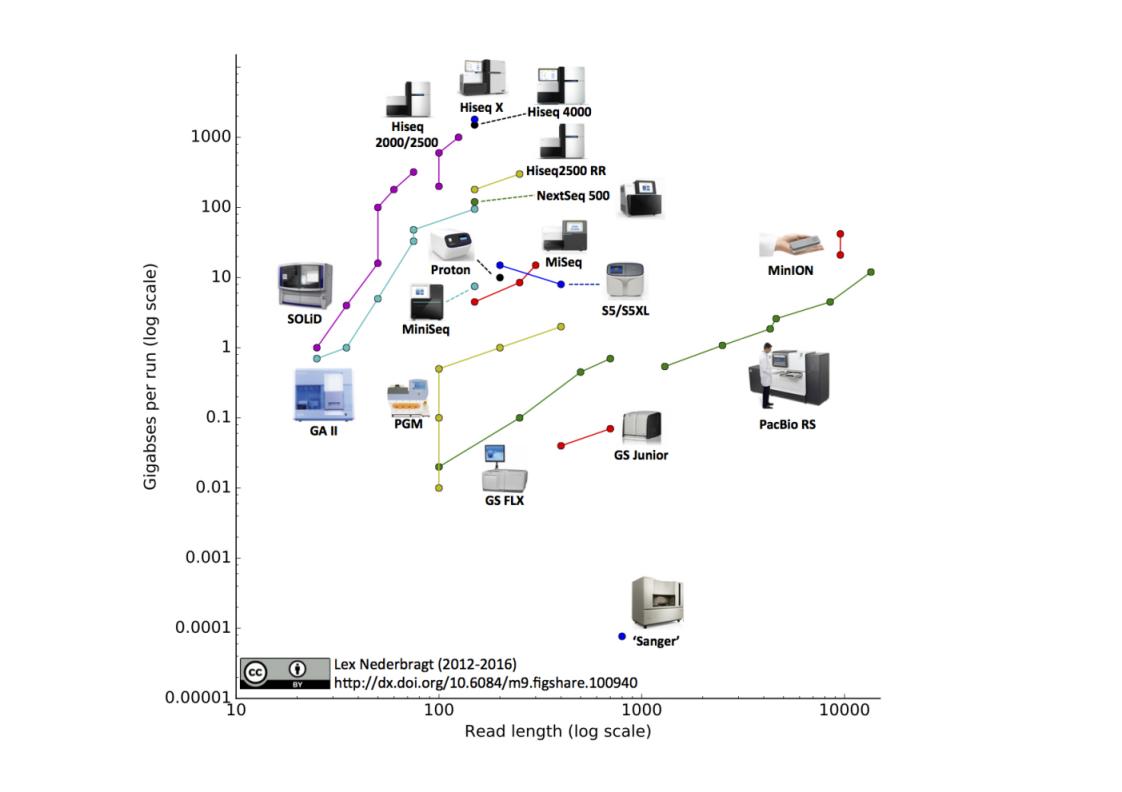
Sanger Sequencing
- Fred Sanger
- 1977
- Uses extension-terminating dideoxynucleotides
- Then
- 1st human genome
- 13 years (1990 - 2003)
- $2.7 billion
Illumina
- Pro
- High throughput, low cost
- Con
- Limited read length hampers complex genome feature (e.g. repeats, low coverage, structural variation) reconstruction in assembly (Partially overcome by paired-end reads with known insert size)
- Takes long time
- Expensive infrastructure
PacBio
- Better assembly
- Lower first pass accuracy
- Faster because sequencing in real time
- Directly detect base modifications
Fastq file
@IL7_1788:5:1:59:769/1
GTGGTCAGTGATTTGCAGGAGGGCACCGGGCCCGTAGATTGCGGCGGCTGGTTAGTGGATGTGTGCGATGCGTTAACCGATCACGCCAGTGAATTTATTGA
+
GGAGAGGG<GGIGIIGIIGGAGGGGGGGGG<AGGGGGGGGGGGGGGGGGGIGGGGGGG<GGGGGIIG<<GAG.AAGGIIIIIGGGAGGGGIGGAGGGGIAG
@IL7_1788:5:1:150:908/18
CCACGCCACAGACCGCTATCAGTCGTCCTTCGCGTATCGCACCCTTAATGTCTTTCATCAGCTGCTTATGGTGGGCAGTTTCATAATACCCGGCCTGTTCA
+
GGGGGGGIIIIIIIGIIGGGIIIGGGGGGGGGGGIGIGGGIIIIIIGGIIIIIIGGGGGIIGIGIIIIGIGGIIIGIIIIIIIGGGGIGGGGGIGGGGGGI

Sequence header
| Sequence header | Meaning |
|---|---|
| @IL7_1788 | instrument name (unique) |
| 5 | flowcell lane |
| 1 | tile number within flow cell |
| 59 | x-coordinate of cluster within tile |
| 769 | y-coordinate of cluster within tile |
| /1 | member of a pair (/1/2) |
Trimming
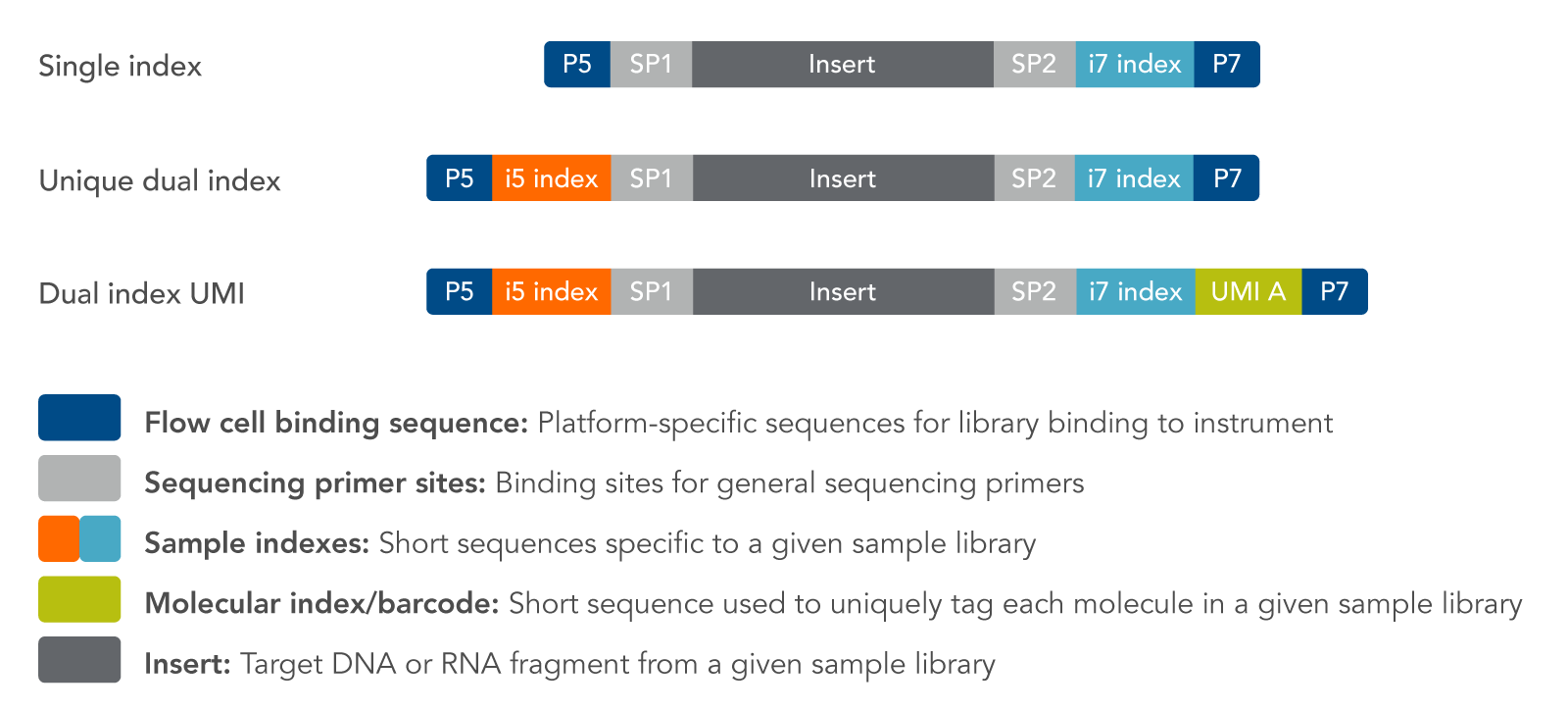
- Need to remove all the adaptors, sequencing primer sites, indices
- Sequencing quality based trimming
https:://sg.idtdna.com/pages/products/next-generation-sequencing/adapters
Workshop
We will do
- View the raw reads file
- Trim the raw reads:
trimmomaticandseqtk - Quality assessment:
fastqc
