05_BLAST
By Yan Li
PhD in Bioinformatics, University of Liverpool
BLAST
- Basic Local Alignment Search Tool
- Compare a query sequence against a database
- Sequence alignment: local & pairwise
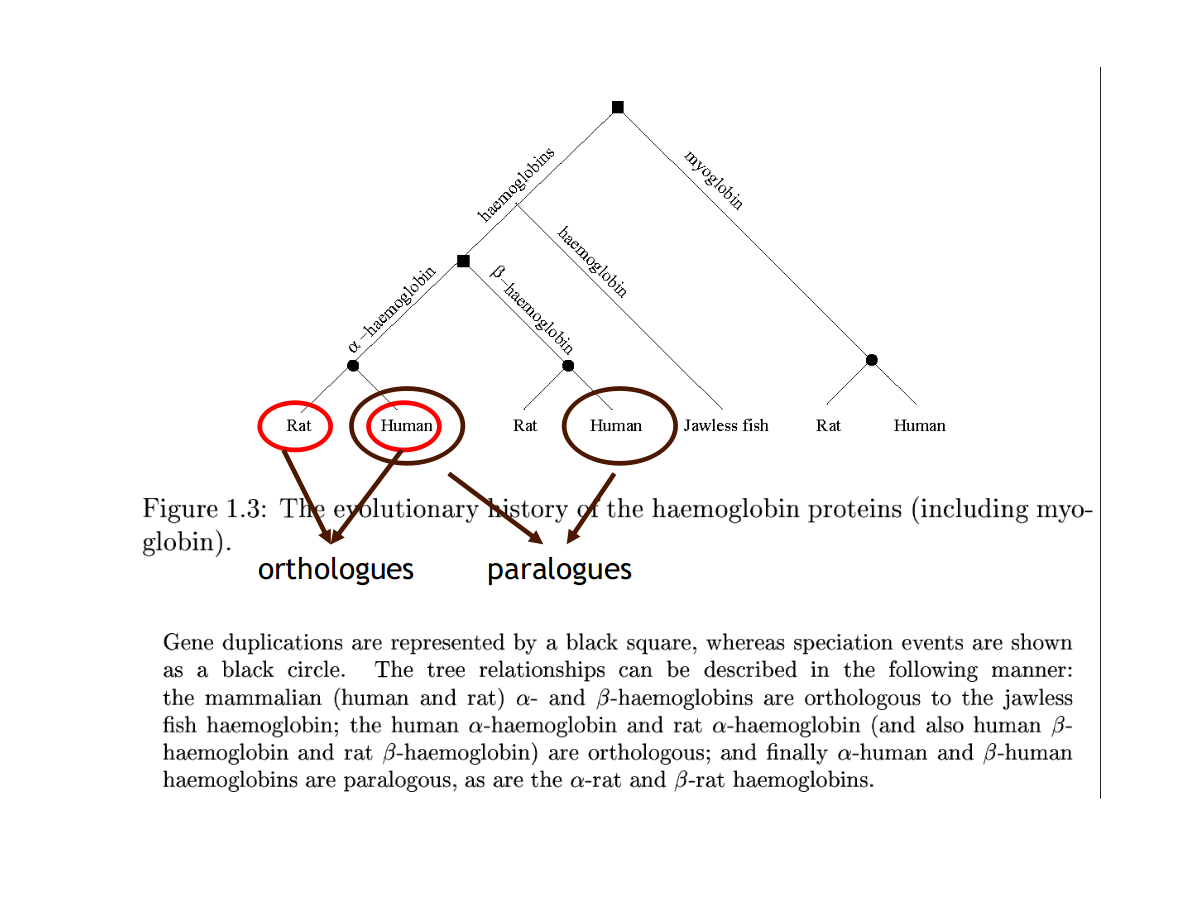
Homology
- Similarity
- Homology: diverged from a common ancestor
- Orthologues: proteins that do the same function in different species
- Paralogues: proteins that perform different, but related functions within one organism
Orthologues vs Paralogues
Blast program
DNA potentially encodes six proteins

BLAST program
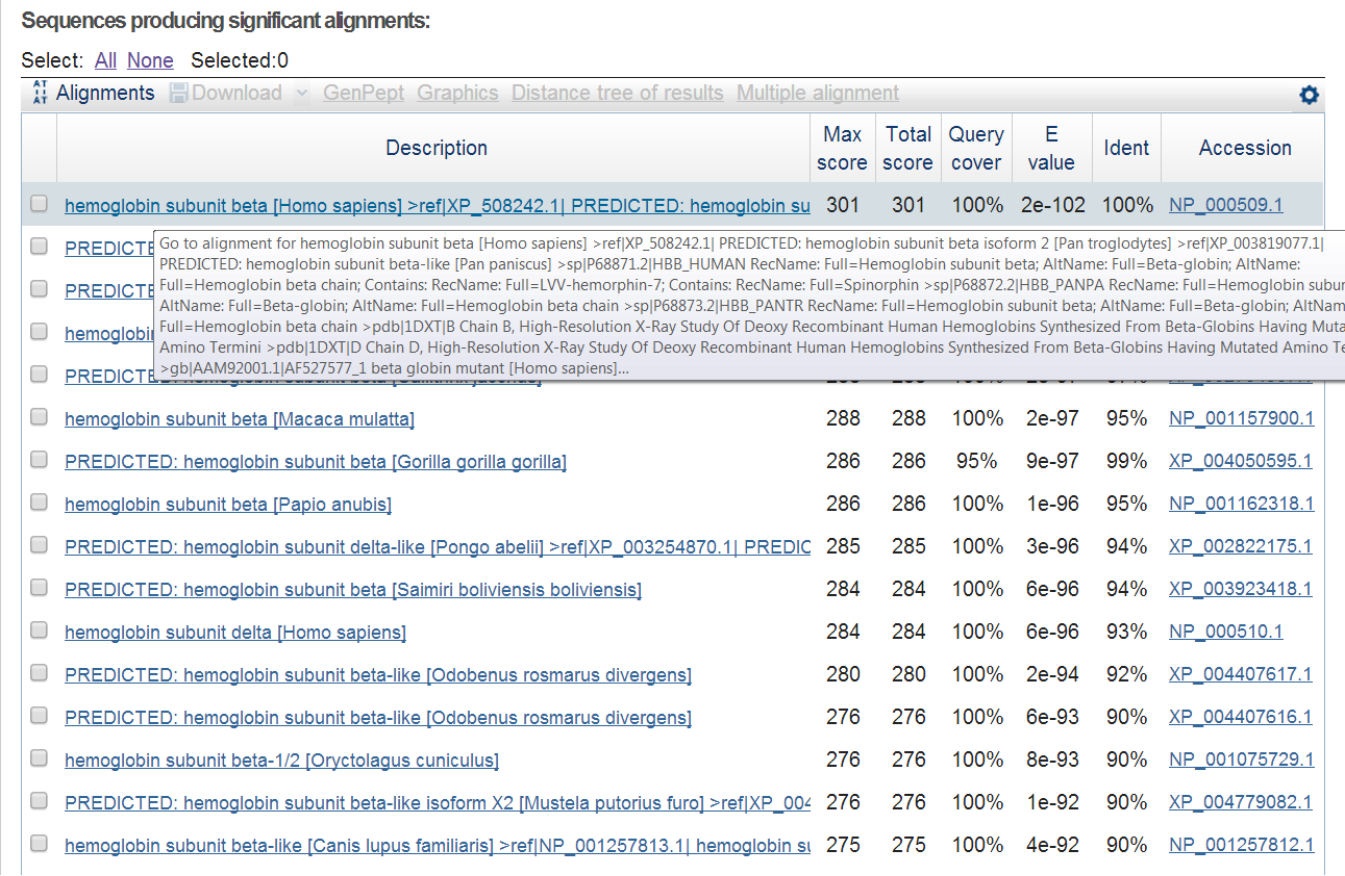
BLAST hit list
BLAST alignment
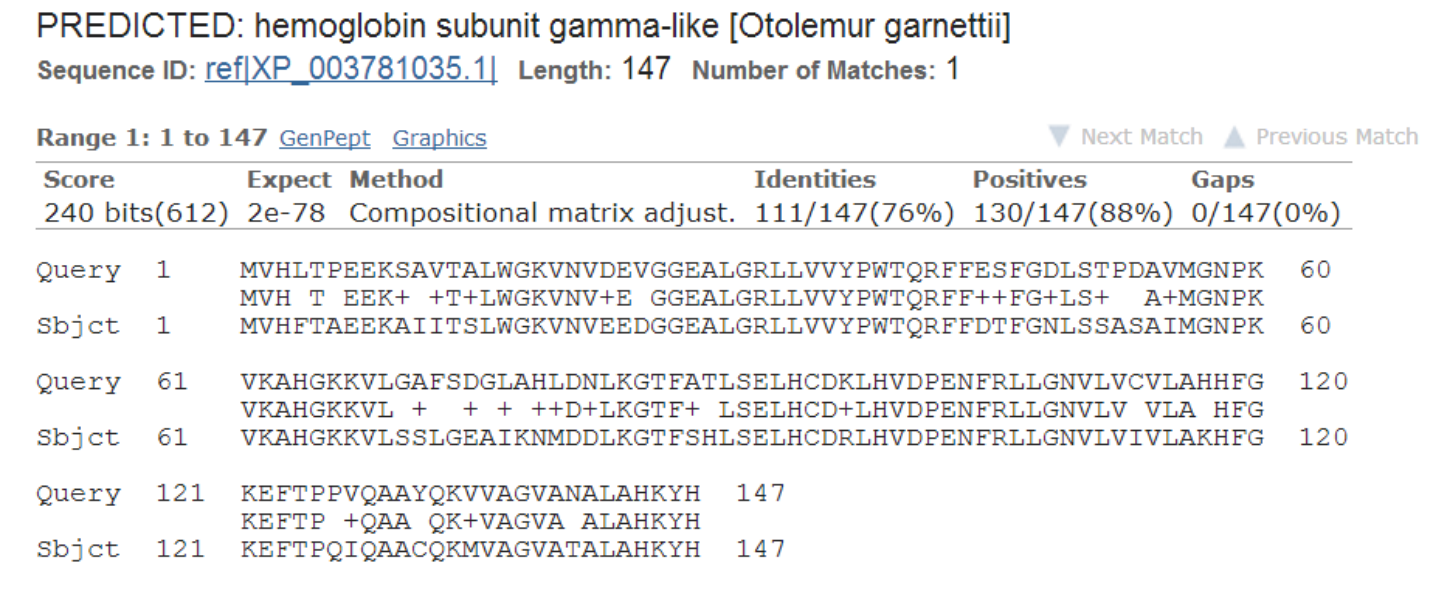
e-value
- Number of matches with this score one can expect to find by chance in a database of size N
- Closer to 0, the better the alignment
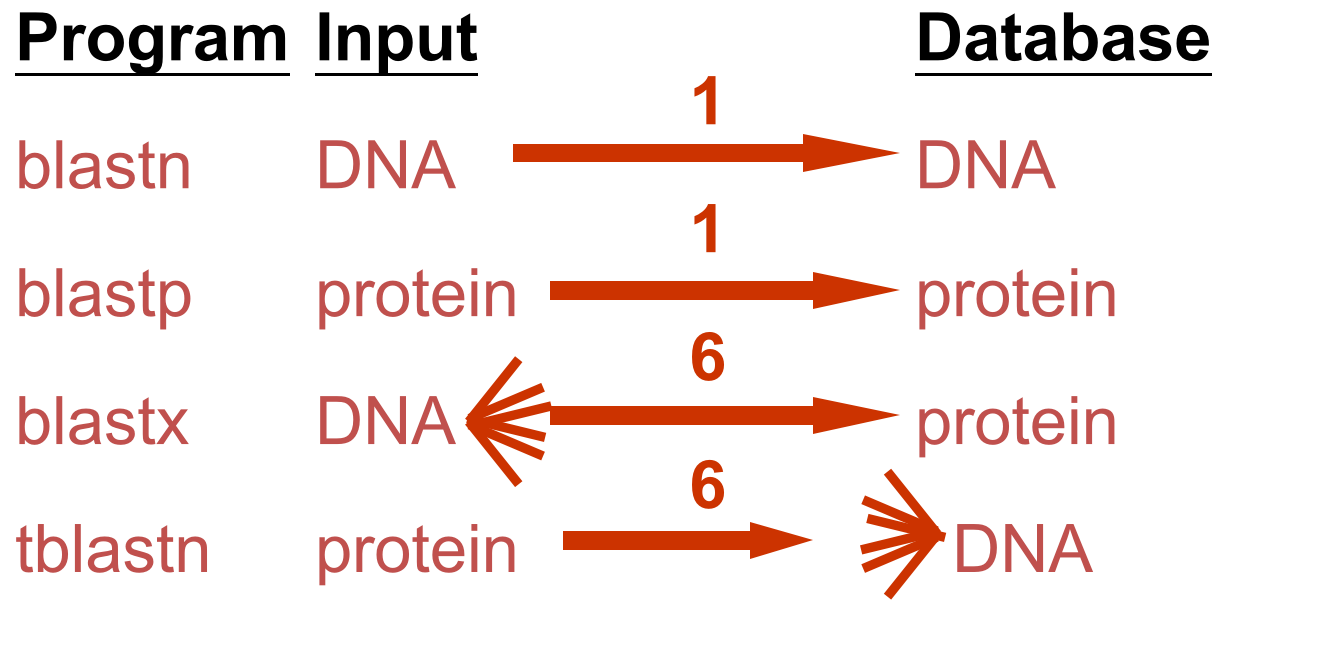
blastn vs blastp
- Faster and more accurate to BLAST proteins rather than nucleotides.
- If you know the reading frame in your sequence – TRANSLATE it and then BLAST
Online BLAST workshop
NCBI (National Center for Biotechnology Information) server:
local BLAST software
conda install -c bioconda blast
makeblastdb -in sopd_gene.fasta -dbtype nucl -out sopD
blastn -query salmonella_typhimurium_lt2.fasta -db sopD -out sopD_lt2.txt -outfmt 1
- Read the help document
- Make notes of all your operations